Introduction¶
Support Vector Machine on Graph Kernels for Protein-Protein Docking Scoring
The software supports the publication of the following articles:
- Geng et al., iScore: A novel graph kernel-based function for scoring protein-protein docking models, bioRxiv 2018, https://doi.org/10.1101/498584
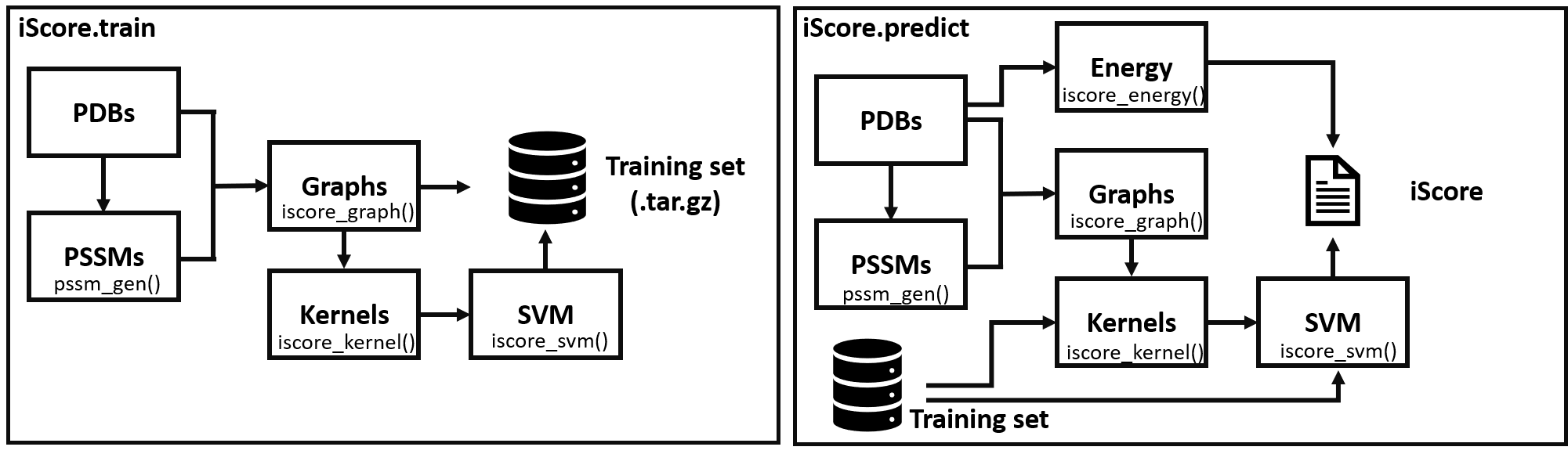
iScore uses a support vector machine (SVM) approach to rank protein-protein interfaces. Each interface is represented by a connection graph in which each node represents a contact residue and each edge the connection between two contact residues of different proterin chain. As feature, the node contains the Position Specific Similarity Matrix (PSSM) of the corresponding residue.
To measure the similarity between two graphs, iScore use a random walk graph kernel (RWGK) approach. These RWGKs are then used as input of the SVM model to either train the model on a training set or use a pretrained model to rank new protein-protein interface.